library(CDMConnector)
requireEunomia()
con <- DBI::dbConnect(duckdb::duckdb(), dbdir = eunomiaDir())
cdm <- cdmFromCon(
con,
cdmSchema = "main",
writeSchema = "main",
writePrefix = "my_study_"
)CohortConstructor
A framework for cohort building in R: the CohortConstructor package for data mapped to the OMOP Common Data Model



The OMOP Common Data Model
Standardising health care data

Standardising health care data

The OMOP CDM tables
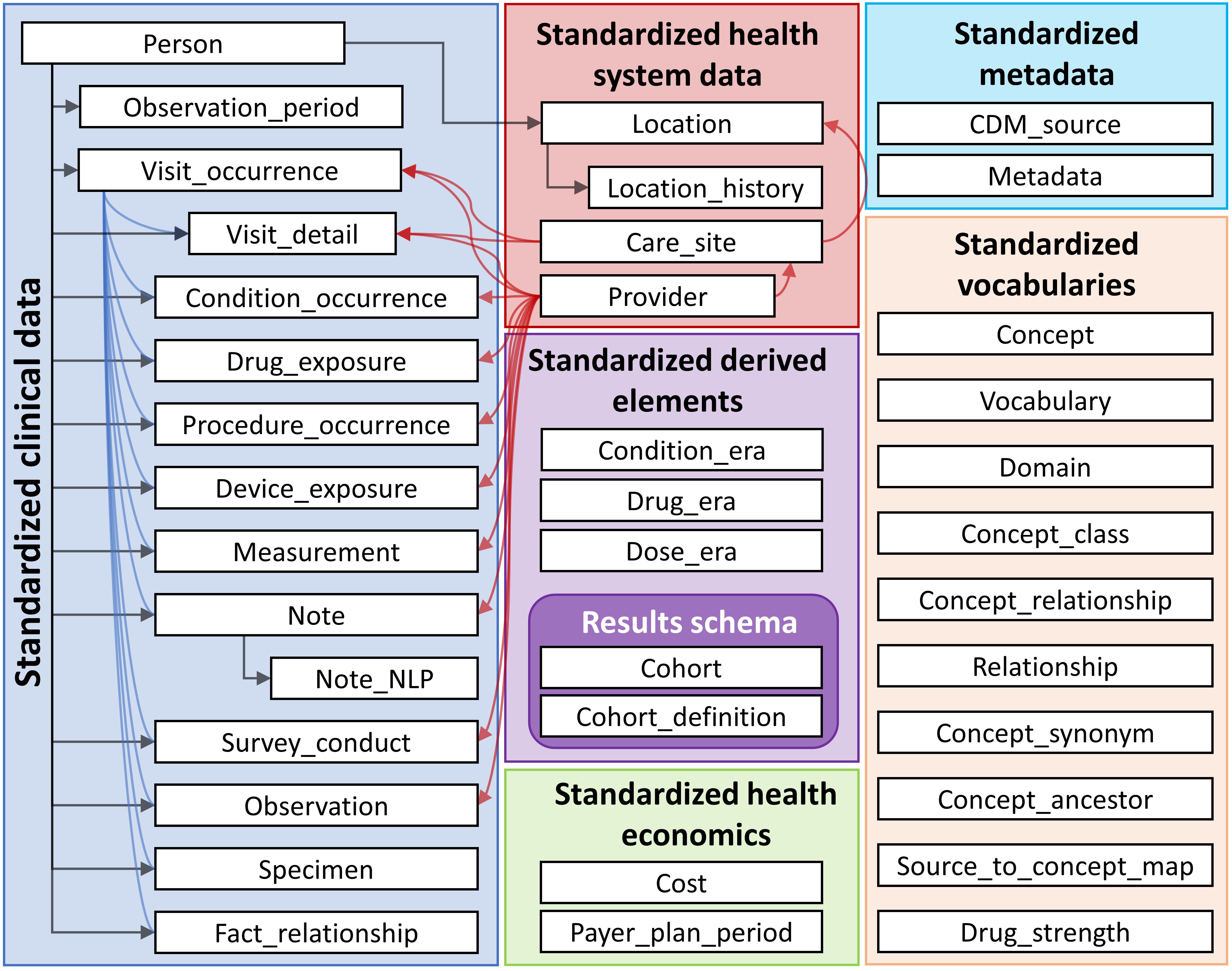
Tables and relation in the OMOP Common Data Model
Creating a reference to the OMOP CDM from R
cdm── # OMOP CDM reference (duckdb) of Synthea ──────────────────────────────────────────────────────────────────────────────────────• omop tables: person, observation_period, visit_occurrence, visit_detail, condition_occurrence, drug_exposure,
procedure_occurrence, device_exposure, measurement, observation, death, note, note_nlp, specimen, fact_relationship, location,
care_site, provider, payer_plan_period, cost, drug_era, dose_era, condition_era, metadata, cdm_source, concept, vocabulary,
domain, concept_class, concept_relationship, relationship, concept_synonym, concept_ancestor, source_to_concept_map,
drug_strength• cohort tables: -• achilles tables: -• other tables: -We’re going to use this example dataset throughout!
Creating a reference to the OMOP CDM from R
Rows: ??
Columns: 18
Database: DuckDB v1.0.0 [eburn@Windows 10 x64:R 4.2.1/C:\Users\eburn\AppData\Local\Temp\RtmpY9GkQd\file7be8112d1fc2.duckdb]
$ person_id <int> 6, 123, 129, 16, 65, 74, 42, 187, 18, 111, 149, 114, 35, 40, 72, 53, 191, 180, 78, 69, 248, …
$ gender_concept_id <int> 8532, 8507, 8507, 8532, 8532, 8532, 8532, 8507, 8532, 8532, 8532, 8532, 8532, 8507, 8532, 85…
$ year_of_birth <int> 1963, 1950, 1974, 1971, 1967, 1972, 1909, 1945, 1965, 1975, 1941, 1972, 1960, 1951, 1947, 19…
$ month_of_birth <int> 12, 4, 10, 10, 3, 1, 11, 7, 11, 5, 8, 3, 3, 12, 7, 8, 6, 4, 1, 10, 8, 6, 7, 6, 11, 7, 2, 3, …
$ day_of_birth <int> 31, 12, 7, 13, 31, 5, 2, 23, 17, 2, 19, 13, 22, 5, 14, 15, 1, 21, 5, 27, 1, 11, 20, 1, 4, 27…
$ birth_datetime <dttm> 1963-12-31, 1950-04-12, 1974-10-07, 1971-10-13, 1967-03-31, 1972-01-05, 1909-11-02, 1945-07…
$ race_concept_id <int> 8516, 8527, 8527, 8527, 8516, 8527, 8527, 8527, 8527, 8527, 8515, 8527, 8527, 8527, 8527, 85…
$ ethnicity_concept_id <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 38003563, 0, 0, 0…
$ location_id <int> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
$ provider_id <int> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
$ care_site_id <int> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
$ person_source_value <chr> "001f4a87-70d0-435c-a4b9-1425f6928d33", "052d9254-80e8-428f-b8b6-69518b0ef3f3", "054d32d5-90…
$ gender_source_value <chr> "F", "M", "M", "F", "F", "F", "F", "M", "F", "F", "F", "F", "F", "M", "F", "M", "F", "F", "M…
$ gender_source_concept_id <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ race_source_value <chr> "black", "white", "white", "white", "black", "white", "white", "white", "white", "white", "a…
$ race_source_concept_id <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ ethnicity_source_value <chr> "west_indian", "italian", "polish", "american", "dominican", "english", "irish", "irish", "e…
$ ethnicity_source_concept_id <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…cdm$person |>
tally()# Source: SQL [?? x 1]
# Database: DuckDB v1.0.0 [eburn@Windows 10 x64:R 4.2.1/C:\Users\eburn\AppData\Local\Temp\RtmpY9GkQd\file7be8112d1fc2.duckdb]
n
<dbl>
1 2694Creating a reference to the OMOP CDM from R
cdm$concept |>
glimpse()Rows: ??
Columns: 10
Database: DuckDB v1.0.0 [eburn@Windows 10 x64:R 4.2.1/C:\Users\eburn\AppData\Local\Temp\RtmpY9GkQd\file7be8112d1fc2.duckdb]
$ concept_id <int> 35208414, 1118088, 40213201, 1557272, 4336464, 4295880, 3020630, 19129655, 44923712, 1569708, 40213216,…
$ concept_name <chr> "Gastrointestinal hemorrhage, unspecified", "celecoxib 200 MG Oral Capsule [Celebrex]", "pneumococcal p…
$ domain_id <chr> "Condition", "Drug", "Drug", "Drug", "Procedure", "Procedure", "Measurement", "Drug", "Drug", "Conditio…
$ vocabulary_id <chr> "ICD10CM", "RxNorm", "CVX", "RxNorm", "SNOMED", "SNOMED", "LOINC", "RxNorm", "NDC", "ICD10CM", "CVX", "…
$ concept_class_id <chr> "4-char billing code", "Branded Drug", "CVX", "Ingredient", "Procedure", "Procedure", "Lab Test", "Clin…
$ standard_concept <chr> NA, "S", "S", "S", "S", "S", "S", "S", NA, NA, "S", "S", "S", "S", "S", "S", NA, "S", "S", "S", "S", "S…
$ concept_code <chr> "K92.2", "213469", "33", "46041", "232717009", "76601001", "2885-2", "789980", "00025152531", "K92", "1…
$ valid_start_date <date> 2007-01-01, 1970-01-01, 2008-12-01, 1970-01-01, 1970-01-01, 1970-01-01, 1970-01-01, 2008-03-30, 2000-0…
$ valid_end_date <date> 2099-12-31, 2099-12-31, 2099-12-31, 2099-12-31, 2099-12-31, 2099-12-31, 2099-12-31, 2099-12-31, 2099-1…
$ invalid_reason <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,…Identifying relevant codes
library(CodelistGenerator)
ingredients <- getDrugIngredientCodes(cdm = cdm)
ingredients
- 10318_tacrine (2 codes)
- 10582_levothyroxine (2 codes)
- 11170_verapamil (2 codes)
- 11248_vitamin_b_12 (2 codes)
- 11289_warfarin (2 codes)
- 11636_drospirenone (2 codes)
along with 85 more codelistsCohorts in OMOP
What Is a Cohort?


A cohort is a set of persons who satisfy one or more inclusion criteria for a duration of time.
Cohorts are defined by sets of clinical codes, and specific logic that defines cohort inclusion, entry and exit.
No distinction between inclusion and exclusion criteria. All criteria are formulated as inclusion criteria.
An individual can contribute to the cohort multiple times, but these cannot overlap. That is, a person can not re-enter the cohort before leaving it.
Individuals must be in observation while contributing time to the cohort.
OMOP Cohorts in R


The
<cohort_table>class is defined in the R packageomopgenerics.This is the class that
CohortConstructoruses, as well as other OMOP analytical packages.-
As defined in
omopgenerics, a<cohort_table>must have at least the following 4 columns (without any missing values in them):cohort_definition_id: Unique identifier for each cohort in the table.
subject_id: Unique patient identifier.
cohort_start_date: Date when the person enters the cohort.
cohort_end_date: Date when the person exits the cohort.
OMOP Cohorts in R


cdm$cohort# Source: table<my_study_cohort> [?? x 4]
# Database: DuckDB v1.0.0 [eburn@Windows 10 x64:R 4.2.1/C:\Users\eburn\AppData\Local\Temp\RtmpY9GkQd\file7be8112d1fc2.duckdb]
cohort_definition_id subject_id cohort_start_date cohort_end_date
<int> <int> <date> <date>
1 1 1177 1980-07-22 1980-08-01
2 1 1478 1969-11-01 1969-11-14
3 1 2747 2008-05-01 2008-05-09
4 1 3567 2010-03-01 2010-03-10
5 1 4027 1986-03-30 1986-04-11
6 1 4081 2015-06-03 2015-06-12
7 1 5017 1988-07-14 1988-07-26
8 1 5113 1971-01-08 1971-01-15
9 1 5329 2009-08-17 2009-08-26
10 2 372 1969-10-05 1969-10-19
# ℹ more rowsOMOP Cohorts in R


Additionally, the <cohort_table> object has the follwing attributes:
- Settings: Relate each cohort definition ID with a cohort name and other variables that define the cohort.
settings(cdm$cohort)# A tibble: 2 × 4
cohort_definition_id cohort_name cdm_version vocabulary_version
<int> <chr> <chr> <chr>
1 1 viral_pharyngitis 5.3 v5.0 18-JAN-19
2 2 viral_sinusitis 5.3 v5.0 18-JAN-19 OMOP Cohorts in R


- Attrition: Store information on each inclusion criteria applied and how many records and subjects were kept after.
attrition(cdm$cohort)# A tibble: 12 × 7
cohort_definition_id number_records number_subjects reason_id reason excluded_records excluded_subjects
<int> <int> <int> <int> <chr> <int> <int>
1 1 10217 2606 1 Initial qualifying events 0 0
2 1 10217 2606 2 Record start <= record end 0 0
3 1 10217 2606 3 Record in observation 0 0
4 1 10217 2606 4 Non-missing sex 0 0
5 1 10217 2606 5 Non-missing year of birth 0 0
6 1 10217 2606 6 Merge overlapping records 0 0
7 2 17268 2686 1 Initial qualifying events 0 0
8 2 17268 2686 2 Record start <= record end 0 0
9 2 17268 2686 3 Record in observation 0 0
10 2 17268 2686 4 Non-missing sex 0 0
11 2 17268 2686 5 Non-missing year of birth 0 0
12 2 17268 2686 6 Merge overlapping records 0 0OMOP Cohorts in R


- Cohort count: Number of records and subjects for each cohort.
cohortCount(cdm$cohort)# A tibble: 2 × 3
cohort_definition_id number_records number_subjects
<int> <int> <int>
1 1 10217 2606
2 2 17268 2686OMOP Cohorts in R


- Cohort codelist: Codelists used to define entry events and inclusion criteria for each cohort.
attr(cdm$cohort, "cohort_codelist")# Source: table<my_study_cohort_codelist> [?? x 4]
# Database: DuckDB v1.0.0 [eburn@Windows 10 x64:R 4.2.1/C:\Users\eburn\AppData\Local\Temp\RtmpY9GkQd\file7be8112d1fc2.duckdb]
cohort_definition_id codelist_name concept_id codelist_type
<int> <chr> <int> <chr>
1 1 viral_pharyngitis 4112343 index event
2 2 viral_sinusitis 40481087 index event CohortConstructor
An R package to build and curate cohorts in the OMOP Common Data Model
Introduction


- CohortConstructor package is designed to support cohort building pipelines in R, using data mapped to the OMOP Common Data Model.
- The code is publicly available in OHDSI’s GitHub repository CohortConstructor.
- CohortConstructor v0.4.0 is available in CRAN.
- Vignettes with further information can be found in the package website.
- More information and context can be found in the online book “Tidy R programming with databases: applications with the OMOP common data model”.
CohortConstructor pipeline


1) Create base cohorts
Cohorts defined using clinical concepts (e.g., asthma diagnoses) or demographics (e.g., females aged >18)
2) Cohort-curation
Tranform base cohorts to meet study-specific inclusion criteria.
Function Sets


Base cohorts Cohort construction based on clinical concepts or demographics.
Requirements and Filtering Demographic restrictions, event presence/absence conditions, and filtering specific records.
Update cohort entry and exit Adjusting entry and exit dates to align with study periods, observation windows, or key events.
Transformation and Combination Merging, stratifying, collapsing, matching, or intersecting cohorts.
Base cohorts
Functions to build base cohorts


Get Started: connecto to Eunomia


# Load relevant packages
library(CDMConnector)
library(CodelistGenerator)
library(CohortConstructor)
library(CohortCharacteristics)
library(dplyr)# Download Eunomia
if (Sys.getenv("EUNOMIA_DATA_FOLDER") == ""){
Sys.setenv("EUNOMIA_DATA_FOLDER" = file.path(tempdir(), "eunomia"))}
if (!dir.exists(Sys.getenv("EUNOMIA_DATA_FOLDER"))){ dir.create(Sys.getenv("EUNOMIA_DATA_FOLDER"))
CDMConnector::downloadEunomiaData()
}
# Connect to the "database"
con <- DBI::dbConnect(duckdb::duckdb(), dbdir = eunomiaDir())
# Create CDM reference object
cdm <- cdmFromCon(
con,
cdmSchema = "main",
writeSchema = "main",
writePrefix = "my_study_"
)Demographics based - Example


- Two cohorts, females and males, both aged 18 to 60 years old, with at least 365 days of previous observation in the database.
cdm$age_cohort <- demographicsCohort(
cdm = cdm,
ageRange = c(18, 60),
sex = c("Female", "Male"),
minPriorObservation = 365,
name = "age_cohort"
)
settings(cdm$age_cohort)# A tibble: 2 × 5
cohort_definition_id cohort_name age_range sex min_prior_observation
<int> <chr> <chr> <chr> <dbl>
1 1 demographics_1 18_60 Female 365
2 2 demographics_2 18_60 Male 365Demographics based - Example


cohortCount(cdm$age_cohort)# A tibble: 2 × 3
cohort_definition_id number_records number_subjects
<int> <int> <int>
1 1 1373 1373
2 2 1321 1321attrition(cdm$age_cohort)# A tibble: 8 × 7
cohort_definition_id number_records number_subjects reason_id reason excluded_records excluded_subjects
<int> <int> <int> <int> <chr> <int> <int>
1 1 2694 2694 1 Initial qualifying events 0 0
2 1 1373 1373 2 Sex requirement: Female 1321 1321
3 1 1373 1373 3 Age requirement: 18 to 60 0 0
4 1 1373 1373 4 Prior observation requirement:… 0 0
5 2 2694 2694 1 Initial qualifying events 0 0
6 2 1321 1321 2 Sex requirement: Male 1373 1373
7 2 1321 1321 3 Age requirement: 18 to 60 0 0
8 2 1321 1321 4 Prior observation requirement:… 0 0Demographics based - Example


To better visualise the attrition, we can use the package CohortCharacteristics to either create a flow diagram or a formatted table:
cdm$age_cohort |> summariseCohortAttrition() |> plotCohortAttrition(type = "png")
Demographics based - Example
cdm$age_cohort |> summariseCohortAttrition() |> tableCohortAttrition()| Reason |
Variable name
|
|||
|---|---|---|---|---|
| number_records | number_subjects | excluded_records | excluded_subjects | |
| Synthea; demographics_1 | ||||
| Initial qualifying events | 2,694 | 2,694 | 0 | 0 |
| Sex requirement: Female | 1,373 | 1,373 | 1,321 | 1,321 |
| Age requirement: 18 to 60 | 1,373 | 1,373 | 0 | 0 |
| Prior observation requirement: 365 days | 1,373 | 1,373 | 0 | 0 |
| Synthea; demographics_2 | ||||
| Initial qualifying events | 2,694 | 2,694 | 0 | 0 |
| Sex requirement: Male | 1,321 | 1,321 | 1,373 | 1,373 |
| Age requirement: 18 to 60 | 1,321 | 1,321 | 0 | 0 |
| Prior observation requirement: 365 days | 1,321 | 1,321 | 0 | 0 |
Concept based - Example


Let’s create a cohort of medications that contains two drugs: diclofenac, and acetaminophen.
- Get relevant codelists with
CodelistGenerator
drug_codes <- getDrugIngredientCodes(
cdm = cdm,
name = c("diclofenac", "acetaminophen"),
nameStyle = "{concept_name}"
)
drug_codes
- acetaminophen (7 codes)
- diclofenac (1 codes)Concept based - Example


- Create concept based cohorts
cdm$medications <- conceptCohort(
cdm = cdm,
conceptSet = drug_codes,
name = "medications"
)
settings(cdm$medications)# A tibble: 2 × 4
cohort_definition_id cohort_name cdm_version vocabulary_version
<int> <chr> <chr> <chr>
1 1 acetaminophen 5.3 v5.0 18-JAN-19
2 2 diclofenac 5.3 v5.0 18-JAN-19 Concept based - Example


- Attrition
| Reason |
Variable name
|
|||
|---|---|---|---|---|
| number_records | number_subjects | excluded_records | excluded_subjects | |
| acetaminophen | ||||
| Initial qualifying events | 14,205 | 2,679 | 0 | 0 |
| Record start <= record end | 14,205 | 2,679 | 0 | 0 |
| Record in observation | 14,205 | 2,679 | 0 | 0 |
| Non-missing sex | 14,205 | 2,679 | 0 | 0 |
| Non-missing year of birth | 14,205 | 2,679 | 0 | 0 |
| Merge overlapping records | 13,908 | 2,679 | 297 | 0 |
| diclofenac | ||||
| Initial qualifying events | 850 | 850 | 0 | 0 |
| Record start <= record end | 850 | 850 | 0 | 0 |
| Record in observation | 830 | 830 | 20 | 20 |
| Non-missing sex | 830 | 830 | 0 | 0 |
| Non-missing year of birth | 830 | 830 | 0 | 0 |
| Merge overlapping records | 830 | 830 | 0 | 0 |
Concept based - Example


- Cohort codelist as an attribute
attr(cdm$medications, "cohort_codelist")# Source: table<my_study_medications_codelist> [?? x 4]
# Database: DuckDB v1.0.0 [eburn@Windows 10 x64:R 4.2.1/C:\Users\eburn\AppData\Local\Temp\RtmpY9GkQd\file7be8604240ee.duckdb]
cohort_definition_id codelist_name concept_id codelist_type
<int> <chr> <int> <chr>
1 1 acetaminophen 1125315 index event
2 1 acetaminophen 1127078 index event
3 1 acetaminophen 1127433 index event
4 1 acetaminophen 40229134 index event
5 1 acetaminophen 40231925 index event
6 1 acetaminophen 40162522 index event
7 1 acetaminophen 19133768 index event
8 2 diclofenac 1124300 index event Requirements and Filtering
Functions to apply requirements and filter


-
On demographics
-
On cohort entries
-
Require presence or absence based on other cohorts, concepts, and tables
Requirement functions - Example


-
We can apply different inclusion and exclusion criteria using CohortConstructor’s functions in a pipe-line fashion. For instance, in what follows we require
only first record per person
subjects 18 years old or more at cohort start date
only females
at least 30 days of prior observation at cohort start date
cdm$medications_requirement <- cdm$medications %>%
requireIsFirstEntry() %>%
requireDemographics(
ageRange = list(c(18, 85)),
sex = "Female",
minPriorObservation = 30,
name = "medications_requirement"
)Requirement functions - Example


result <- cdm$medications_requirement |>
summariseCohortAttrition(cohortId = 1)
result |>
tableCohortAttrition(
groupColumn = c("cohort_name"),
hide = c("variable_level", "reason_id", "estimate_name", "cdm_name", settingsColumns(result))
)| Reason |
Variable name
|
|||
|---|---|---|---|---|
| number_records | number_subjects | excluded_records | excluded_subjects | |
| acetaminophen | ||||
| Initial qualifying events | 14,205 | 2,679 | 0 | 0 |
| Record start <= record end | 14,205 | 2,679 | 0 | 0 |
| Record in observation | 14,205 | 2,679 | 0 | 0 |
| Non-missing sex | 14,205 | 2,679 | 0 | 0 |
| Non-missing year of birth | 14,205 | 2,679 | 0 | 0 |
| Merge overlapping records | 13,908 | 2,679 | 297 | 0 |
| Restricted to first entry | 2,679 | 2,679 | 11,229 | 0 |
| Age requirement: 18 to 85 | 308 | 308 | 2,371 | 2,371 |
| Sex requirement: Female | 175 | 175 | 133 | 133 |
| Prior observation requirement: 30 days | 175 | 175 | 0 | 0 |
| Future observation requirement: 0 days | 175 | 175 | 0 | 0 |
Update cohort entry and exit
Functions to update cohort start and end dates


-
Cohort entry
-
Trim start and end dates
-
Pad start and end dates
Update cohort entry and exit - Example


We can trim start and end dates to match demographic requirements.
-
For instance cohort dates can be trimmed so the subject contributes time while:
Aged 20 to 40 years old
Prior observation of at least 365 days
cdm$medications_trimmed <- cdm$medications %>%
trimDemographics(
ageRange = list(c(20, 40)),
minPriorObservation = 365,
name = "medications_trimmed"
)Update cohort entry and exit - Example


result <- cdm$medications_trimmed |>
summariseCohortAttrition(cohortId = 1)
result |>
tableCohortAttrition(
groupColumn = c("cohort_name"),
hide = c("variable_level", "reason_id", "estimate_name", "cdm_name", settingsColumns(result))
)| Reason |
Variable name
|
|||
|---|---|---|---|---|
| number_records | number_subjects | excluded_records | excluded_subjects | |
| acetaminophen | ||||
| Initial qualifying events | 14,205 | 2,679 | 0 | 0 |
| Record start <= record end | 14,205 | 2,679 | 0 | 0 |
| Record in observation | 14,205 | 2,679 | 0 | 0 |
| Non-missing sex | 14,205 | 2,679 | 0 | 0 |
| Non-missing year of birth | 14,205 | 2,679 | 0 | 0 |
| Merge overlapping records | 13,908 | 2,679 | 297 | 0 |
| Restricted to first entry | 2,679 | 2,679 | 11,229 | 0 |
| Age requirement: 20 to 40 | 222 | 222 | 2,457 | 2,457 |
| Prior observation requirement: 365 days | 222 | 222 | 0 | 0 |
Transformation and Combination
Functions for Cohort Transformation and Combination


-
Split cohorts
-
Combine cohorts
-
Filter cohorts
-
Match cohorts
-
Concatenate entries
-
Copy and rename cohorts
Cohort combinations - Example


Collapse entries of acetaminophen and diclofenac, so if the gap is 7 days or less, entries are merged.
Create a new cohort that contains people who had an exposure to both diclofenac and acetaminophen at the same time using.
cdm$intersection <- cdm$medications |>
collapseCohorts(gap = 7) |>
CohortConstructor::intersectCohorts(
gap = 7,
name = "intersection"
)
settings(cdm$intersection)# A tibble: 1 × 5
cohort_definition_id cohort_name gap acetaminophen diclofenac
<int> <chr> <dbl> <dbl> <dbl>
1 1 acetaminophen_diclofenac 7 1 1Cohort combinations - Example


attr(cdm$intersection, "cohort_codelist")# Source: table<my_study_intersection_codelist> [?? x 4]
# Database: DuckDB v1.0.0 [eburn@Windows 10 x64:R 4.2.1/C:\Users\eburn\AppData\Local\Temp\RtmpY9GkQd\file7be8604240ee.duckdb]
cohort_definition_id codelist_name concept_id codelist_type
<int> <chr> <int> <chr>
1 1 acetaminophen 1125315 index event
2 1 acetaminophen 1127078 index event
3 1 acetaminophen 1127433 index event
4 1 acetaminophen 40229134 index event
5 1 acetaminophen 40231925 index event
6 1 acetaminophen 40162522 index event
7 1 acetaminophen 19133768 index event
8 1 diclofenac 1124300 index event PhenotypeR
An R package to assess the research-readiness of a set of cohorts in the OMOP Common Data Model
Diagnostics
Database diagnostics: Information to understand the database where the cohorts have been created.
Codelists diagnostics: Which of the concepts are used in the database, and in the cohorts? In which frequency? Are we missing any codes?
Cohort diagnostics: How many people are in the cohorts? Which was the impact of the inclusion criteria? Which are the characteristics of the patients in the cohorts?
Matched diagnostics: Compare characteristics of the people in the cohorts to matched pairs (sex and age) in the general database population.
Population diagnostics: Incidence and Prevalence of the cohorts in the database.
Functions
-
Run all diagnostics
phenotypeDiagnostics()
-
Run individual diagnostics
codelistDiagnostics()cohortDiagnostics()databaseDiagnostics()matchedDiagnostics()populationDiagnostics()
-
Visualise results
shinyDiagnostics()
Example
We can easily run all the diagnostics explain as follows:
result <- phenotypeDiagnostics(cdm$medications)Once we have results, we can creat interactive application to revise results.
shinyDiagnostics(result = result, directory = tempdir())See an example shiny app here.
ED PART 2
Thank you!
Questions?



Exercises
Exercise 1 - Base Cohorts
Create a cohort of aspirin use. Consider that two records separated by less than 1 week, can be considered as a continuous exposure.
- How many records does it have? And how many subjects?
| CDM name | Variable name | Estimate name |
Cohort name
|
|---|---|---|---|
| aspirin | |||
| Synthea | Number records | N | 4,379 |
| Number subjects | N | 1,927 |
Exercise 2 - Requirement and filtering
Create a new cohort named “aspirin_last” by applying the following criteria to the base aspirin cohort:
Include only the last drug exposure for each subject.
Include exposures that start between January 1, 1960, and December 31, 1979.
Exclude individuals with an amoxicillin exposure in the 7 days prior to the aspirin exposure.
Move to the next slide to see the attrition.
Exercise 2 - Requirement and filtering
| Reason |
Variable name
|
|||
|---|---|---|---|---|
| number_records | number_subjects | excluded_records | excluded_subjects | |
| Synthea; aspirin | ||||
| Initial qualifying events | 4,380 | 1,927 | 0 | 0 |
| Record start <= record end | 4,380 | 1,927 | 0 | 0 |
| Record in observation | 4,380 | 1,927 | 0 | 0 |
| Non-missing sex | 4,380 | 1,927 | 0 | 0 |
| Non-missing year of birth | 4,380 | 1,927 | 0 | 0 |
| Merge overlapping records | 4,379 | 1,927 | 1 | 0 |
| Restricted to last entry | 1,927 | 1,927 | 2,452 | 0 |
| cohort_start_date after 1960-01-01 | 1,511 | 1,511 | 416 | 416 |
| cohort_start_date before 1979-12-31 | 1,174 | 1,174 | 337 | 337 |
| Not in concept amoxicillin between -7 & 0 days relative to cohort_start_date | 1,163 | 1,163 | 11 | 11 |
Exercise 3 - Update cohort entry and exit
Create a cohort of ibuprofen. From it, create an “ibuprofen_death” cohort which includes only subjects that have a future record of death in the database, and update cohort end date to be the death date.
| Reason |
Variable name
|
|||
|---|---|---|---|---|
| number_records | number_subjects | excluded_records | excluded_subjects | |
| Synthea; ibuprofen | ||||
| Initial qualifying events | 2,148 | 1,451 | 0 | 0 |
| Record start <= record end | 2,148 | 1,451 | 0 | 0 |
| Record in observation | 2,148 | 1,451 | 0 | 0 |
| Non-missing sex | 2,148 | 1,451 | 0 | 0 |
| Non-missing year of birth | 2,148 | 1,451 | 0 | 0 |
| Merge overlapping records | 2,148 | 1,451 | 0 | 0 |
| No death recorded | 0 | 0 | 2,148 | 1,451 |
| Exit at death | 0 | 0 | 0 | 0 |
Exercise 4 - Transformation and Combination
From the ibuprofen base cohort (not subseted to death), create five separate cohorts. Each cohort should include records for one specific year from the following list: 1975, 1976, 1977, 1978, 1979, and 1980.
- How many records and subjects are in each cohort?
| CDM name | Variable name | Estimate name |
Cohort name
|
|||||
|---|---|---|---|---|---|---|---|---|
| ibuprofen_1975 | ibuprofen_1976 | ibuprofen_1977 | ibuprofen_1978 | ibuprofen_1979 | ibuprofen_1980 | |||
| Synthea | Number records | N | 71 | 64 | 60 | 75 | 66 | 63 |
| Number subjects | N | 68 | 61 | 60 | 74 | 66 | 63 | |
Exercise 5
Use CohortConstructor to create a cohort with the following criteria:
Users of diclofenac
Females aged 16 or older
With at least 365 days of continuous observation prior to exposure
Without prior exposure to any of amoxicillin
With cohort exit defined as first discontinuation of exposure. An exposure being define as recorded exposures within 7-days gap.
Exercise 5
| Reason |
Variable name
|
|||
|---|---|---|---|---|
| number_records | number_subjects | excluded_records | excluded_subjects | |
| Synthea; diclofenac | ||||
| Initial qualifying events | 850 | 850 | 0 | 0 |
| Record start <= record end | 850 | 850 | 0 | 0 |
| Record in observation | 830 | 830 | 20 | 20 |
| Non-missing sex | 830 | 830 | 0 | 0 |
| Non-missing year of birth | 830 | 830 | 0 | 0 |
| Merge overlapping records | 830 | 830 | 0 | 0 |
| Age requirement: 16 to 150 | 830 | 830 | 0 | 0 |
| Sex requirement: Female | 435 | 435 | 395 | 395 |
| Prior observation requirement: 365 days | 435 | 435 | 0 | 0 |
| Future observation requirement: 0 days | 435 | 435 | 0 | 0 |
| Not in concept amoxicillin between -Inf & -1 days relative to cohort_start_date | 161 | 161 | 274 | 274 |
| Collapse cohort with a gap of 7 days. | 161 | 161 | 0 | 0 |
| Restricted to first entry | 161 | 161 | 0 | 0 |
R/Medicine 2025