library(DBI)
library(here)
library(dplyr)2 Creating a reference to data in the OMOP CDM
An entity-relationship diagram for version 5.4 of the OMOP CDM is shown below. 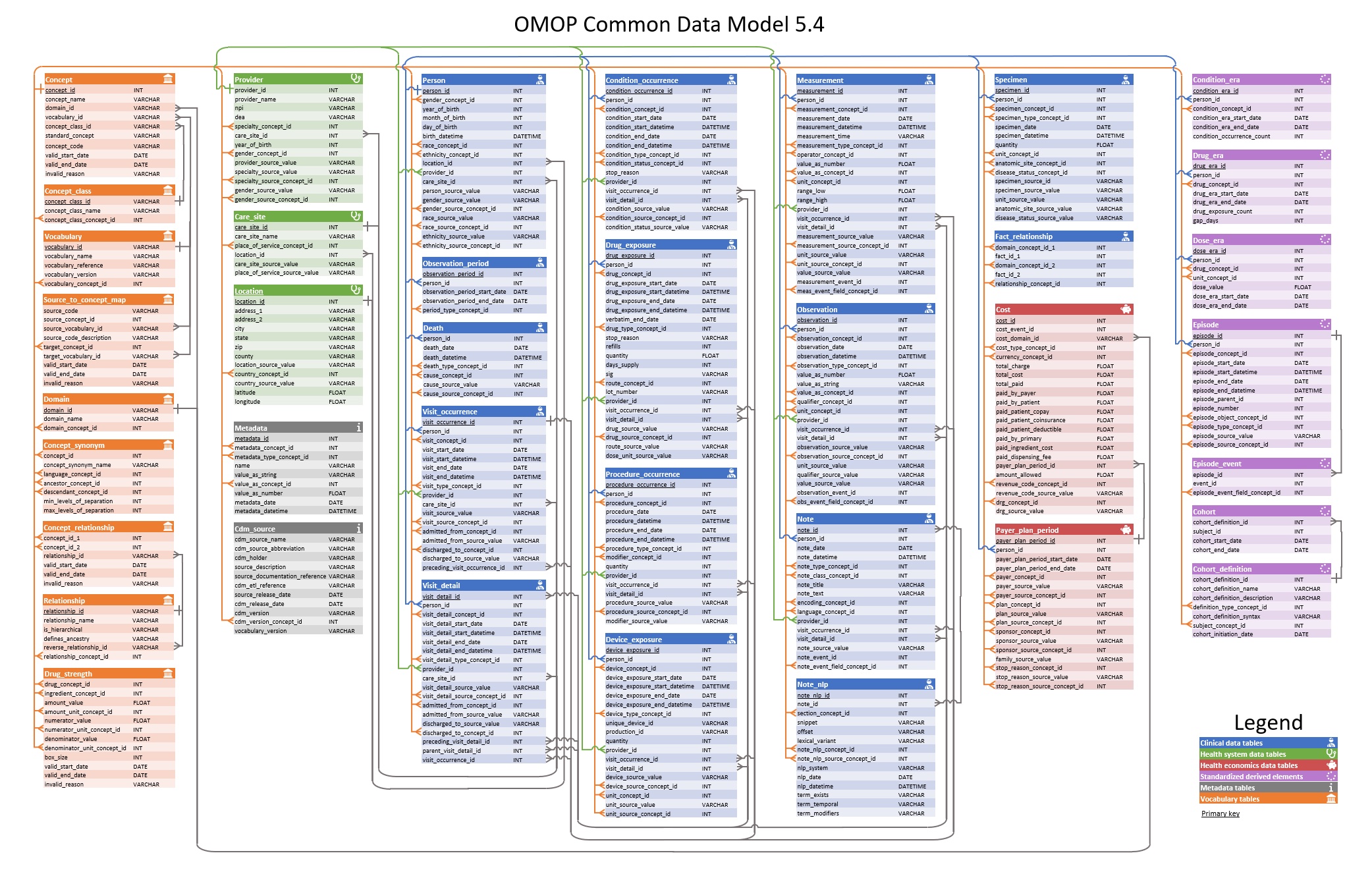
2.1 Connecting to a database from R using DBI
Database connections from R can be made using the DBI package. The back-end for DBI is facilitated by database specific driver packages. We saw in Chapter 1 an example where we created a new, empty, in-process duckdb database which we then added database. But we could have instead connected to an existing duckdb database. This could, for example, look like
db <- dbConnect(duckdb::duckdb(),
dbdir = here("my_duckdb_database.ducdkb"))For other database management systems, creating connections would be supported by the associated back-end packages. For example a connection to a Postgres database would look something like:
db <- DBI::dbConnect(RPostgres::Postgres(),
dbname = Sys.getenv("CDM5_POSTGRESQL_DBNAME"),
host = Sys.getenv("CDM5_POSTGRESQL_HOST"),
user = Sys.getenv("CDM5_POSTGRESQL_USER"),
password = Sys.getenv("CDM5_POSTGRESQL_PASSWORD"))2.2 Referencing tables from the OMOP common data model
As seen in the previous chapter, once a connection to the database has been created we can then create references to the various tables in the database and build queries using in a familiar dplyr style. To show this, let’s download an example dataset (synthea-covid19-10k) provided by CDMConnector.
Now we have this downloaded, we can create a connection to a duckdb database containing the data in a similar way to how we’ve done before.
db <- dbConnect(
duckdb::duckdb(),
dbdir = CDMConnector::eunomiaDir(datasetName = "synthea-covid19-10k"))
db |>
tbl("person")# Source: table<person> [?? x 18]
# Database: DuckDB v1.1.3 [eburn@Windows 10 x64:R 4.4.0/C:\Users\eburn\AppData\Local\Temp\Rtmpoz0O3N\file54bc5ba63902.duckdb]
person_id gender_concept_id year_of_birth month_of_birth day_of_birth
<int> <int> <int> <int> <int>
1 1 8532 1970 4 24
2 2 8532 1929 3 18
3 3 8532 1970 4 4
4 4 8507 1966 2 26
5 5 8532 1936 6 10
6 6 8507 1996 5 29
7 7 8507 1923 11 14
8 8 8507 2018 8 20
9 9 8532 1933 2 11
10 10 8507 2010 3 7
# ℹ more rows
# ℹ 13 more variables: birth_datetime <dttm>, race_concept_id <int>,
# ethnicity_concept_id <int>, location_id <int>, provider_id <int>,
# care_site_id <int>, person_source_value <chr>, gender_source_value <chr>,
# gender_source_concept_id <int>, race_source_value <chr>,
# race_source_concept_id <int>, ethnicity_source_value <chr>,
# ethnicity_source_concept_id <int>db |>
tbl("observation_period")# Source: table<observation_period> [?? x 5]
# Database: DuckDB v1.1.3 [eburn@Windows 10 x64:R 4.4.0/C:\Users\eburn\AppData\Local\Temp\Rtmpoz0O3N\file54bc5ba63902.duckdb]
observation_period_id person_id observation_period_s…¹ observation_period_e…²
<int> <int> <date> <date>
1 1 1 2014-05-09 2023-05-12
2 2 2 1977-04-11 1986-09-15
3 3 3 2014-04-19 2023-04-22
4 4 4 2014-03-22 2023-04-08
5 5 5 2013-11-13 2023-01-04
6 6 6 2013-07-17 2021-08-04
7 7 7 2013-06-26 2022-08-17
8 8 8 2018-08-20 2022-07-25
9 9 9 2013-08-03 2022-09-24
10 10 10 2013-08-11 2023-04-02
# ℹ more rows
# ℹ abbreviated names: ¹observation_period_start_date,
# ²observation_period_end_date
# ℹ 1 more variable: period_type_concept_id <int>We could also perform similar queries to those seen in chapter 1, but this time working with the patient-level, rather than penguin-level, data.
db |>
tbl("person") |>
group_by(year_of_birth) |>
count() # Source: SQL [?? x 2]
# Database: DuckDB v1.1.3 [eburn@Windows 10 x64:R 4.4.0/C:\Users\eburn\AppData\Local\Temp\Rtmpoz0O3N\file54bc5ba63902.duckdb]
# Groups: year_of_birth
year_of_birth n
<int> <dbl>
1 1923 66
2 1924 123
3 1925 140
4 1926 146
5 1927 162
6 1928 136
7 1929 146
8 1930 119
9 1931 135
10 1932 142
# ℹ more rows2.3 Creating a reference to the OMOP common data model
As the structure of the OMOP CDM is already known, we can avoid the overhead of creating individual references to the OMOP CDM tables like above by instead creating a joint reference for all OMOP CDM database tables in one go.
The R object representing OMOP CDM data is defined by the omopgenerics package), with the the CDMConnector package providing a means of connecting to a OMOP CDM data held in a database. As well as specifying the schema containing our OMOP CDM tables, we also specify a write schema where any database tables we create during our analysis will be stored (often our OMOP CDM tables will be in a schema that we only have read-access to and we’ll have another schema where we can have write-access where we intermediate tables are created for a given a study).
library(omopgenerics)Warning: package 'omopgenerics' was built under R version 4.4.2
Attaching package: 'omopgenerics'The following object is masked from 'package:stats':
filterlibrary(CDMConnector)
Attaching package: 'CDMConnector'The following objects are masked from 'package:omopgenerics':
cdmName, recordCohortAttrition, uniqueTableNamecdm <- cdmFromCon(con = db,
cdmSchema = "main",
writeSchema = "main",
cdmName = "Synthea Covid-19 data")Note: method with signature 'DBIConnection#Id' chosen for function 'dbExistsTable',
target signature 'duckdb_connection#Id'.
"duckdb_connection#ANY" would also be validcdm── # OMOP CDM reference (duckdb) of Synthea Covid-19 data ──────────────────────• omop tables: person, observation_period, visit_occurrence, visit_detail,
condition_occurrence, drug_exposure, procedure_occurrence, device_exposure,
measurement, observation, death, note, note_nlp, specimen, fact_relationship,
location, care_site, provider, payer_plan_period, cost, drug_era, dose_era,
condition_era, metadata, cdm_source, concept, vocabulary, domain,
concept_class, concept_relationship, relationship, concept_synonym,
concept_ancestor, source_to_concept_map, drug_strength, cohort_definition,
attribute_definition• cohort tables: -• achilles tables: -• other tables: -We can also specify a write prefix and this will be used whenever permanent tables are created the write schema. This can be useful when we’re sharing our write schema with others and want to avoid table name conflicts and easily drop tables created as part of a particular study.
cdm <- cdmFromCon(con = db,
cdmSchema = "main",
writeSchema = "main",
writePrefix = "my_study_",
cdmName = "Synthea Covid-19 data")We can see that we now have an object that contains references to all the OMOP CDM tables. We can reference specific tables using the “$” or “[[ … ]]” operators.
cdm$person# Source: table<main.person> [?? x 18]
# Database: DuckDB v1.1.3 [eburn@Windows 10 x64:R 4.4.0/C:\Users\eburn\AppData\Local\Temp\Rtmpoz0O3N\file54bc5ba63902.duckdb]
person_id gender_concept_id year_of_birth month_of_birth day_of_birth
<int> <int> <int> <int> <int>
1 1 8532 1970 4 24
2 2 8532 1929 3 18
3 3 8532 1970 4 4
4 4 8507 1966 2 26
5 5 8532 1936 6 10
6 6 8507 1996 5 29
7 7 8507 1923 11 14
8 8 8507 2018 8 20
9 9 8532 1933 2 11
10 10 8507 2010 3 7
# ℹ more rows
# ℹ 13 more variables: birth_datetime <dttm>, race_concept_id <int>,
# ethnicity_concept_id <int>, location_id <int>, provider_id <int>,
# care_site_id <int>, person_source_value <chr>, gender_source_value <chr>,
# gender_source_concept_id <int>, race_source_value <chr>,
# race_source_concept_id <int>, ethnicity_source_value <chr>,
# ethnicity_source_concept_id <int>cdm[["observation_period"]]# Source: table<main.observation_period> [?? x 5]
# Database: DuckDB v1.1.3 [eburn@Windows 10 x64:R 4.4.0/C:\Users\eburn\AppData\Local\Temp\Rtmpoz0O3N\file54bc5ba63902.duckdb]
observation_period_id person_id observation_period_s…¹ observation_period_e…²
<int> <int> <date> <date>
1 1 1 2014-05-09 2023-05-12
2 2 2 1977-04-11 1986-09-15
3 3 3 2014-04-19 2023-04-22
4 4 4 2014-03-22 2023-04-08
5 5 5 2013-11-13 2023-01-04
6 6 6 2013-07-17 2021-08-04
7 7 7 2013-06-26 2022-08-17
8 8 8 2018-08-20 2022-07-25
9 9 9 2013-08-03 2022-09-24
10 10 10 2013-08-11 2023-04-02
# ℹ more rows
# ℹ abbreviated names: ¹observation_period_start_date,
# ²observation_period_end_date
# ℹ 1 more variable: period_type_concept_id <int>2.4 CDM attributes
2.4.1 CDM name
Our cdm reference will be associated with a name. By default this name will be taken from the cdm source name field from the cdm source table. We can though set this to a different name when creating our cdm reference. This cdm name attribute of our reference is particularly useful in the context of network studies to keep track of which results are associated with which database.
cdm <- cdmFromCon(db,
cdmSchema = "main",
writeSchema = "main")
cdm$cdm_source# Source: table<main.cdm_source> [?? x 10]
# Database: DuckDB v1.1.3 [eburn@Windows 10 x64:R 4.4.0/C:\Users\eburn\AppData\Local\Temp\Rtmpoz0O3N\file54bc5ba63902.duckdb]
cdm_source_name cdm_source_abbreviation cdm_holder source_description
<chr> <chr> <chr> <chr>
1 Synthea Synthea "" Synthea Synthetic Data
# ℹ 6 more variables: source_documentation_reference <chr>,
# cdm_etl_reference <chr>, source_release_date <date>,
# cdm_release_date <date>, cdm_version <chr>, vocabulary_version <chr>cdmName(cdm)[1] "Synthea"cdm <- cdmFromCon(db,
cdmSchema = "main",
writeSchema = "main",
cdmName = "my_cdm")
cdmName(cdm)[1] "my_cdm"2.4.2 CDM version
We can also easily check the OMOP CDM version that is being used
cdmVersion(cdm)[1] "5.3"2.4.3 CDM Source
Although typically we won’t need to use them for writing study code, we can also access lower-level information on the source, such as the database connection.
attr(cdmSource(cdm), "dbcon")<duckdb_connection c6560 driver=<duckdb_driver dbdir='C:\Users\eburn\AppData\Local\Temp\Rtmpoz0O3N\file54bc5ba63902.duckdb' read_only=FALSE bigint=numeric>>3 Disconnecting
Once we have finished our analysis we can close our connection to the database behind our cdm reference like so.
cdmDisconnect(cdm)